Example 2: Start from nifti files#
This is similar to when processing is started from dicom-files. The only differece is that the result folder is already present with the necessary files. It should look like this:
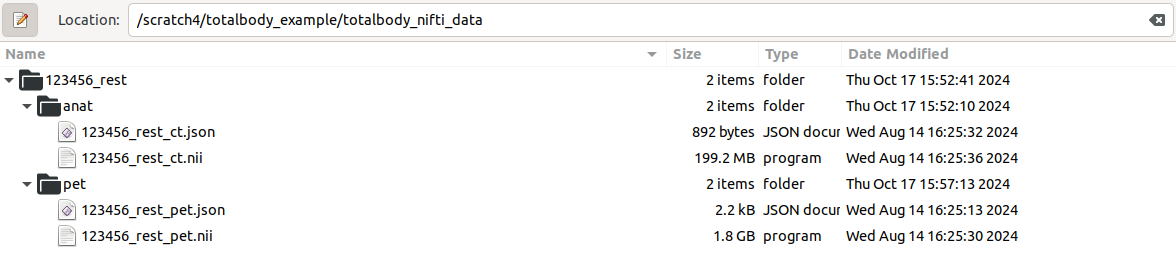
The json-files are generated when dicoms are converted to nifti using dcm2niix. The json contains header information from dicom which means the study information doesn’t need to be manually fetched.
To process this subject, type:
run_totalbody_turbo('123456_rest')
This is works exactly the same as starting from dicom files. Result should look like this:
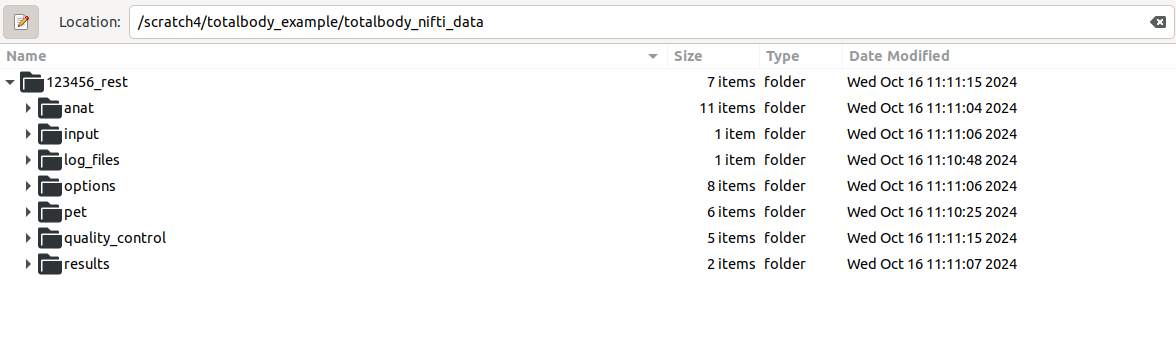
If json files are not present, study_specs struct needs to be specified by the user.
custom_study_specs = struct(
frames = [frame_start, frame_end], … % Nx2 matrix
tracer = '[15o]h2o', ...
dose = 398, …
weight = 92, ...
dc = 1, ...
mri_code = '0', ...
glucose = NaN, … % fdg only
hct = NaN, … % fdg only
)
run_totalbody_turbo('123456_rest', study_specs = custom_study_specs)
To create a struct with all necessary fields, you can also use set_study_specs. This initializes a struct with NaN and empty values that you can set yourself.
custom_study_specs = set_study_specs();
custom_study_specs(...
frames = [frame_start, frame_end], … Nx2 matrix
tracer = '[15o]h2o', ...
dose = 398, …
weight = 92, ...
dc = 1, ...
mri_code = '0', ...
glucose = NaN, … % fdg only
hct = NaN, … % fdg only
)
run_totalbody_turbo('123456_rest', study_specs = custom_study_specs)
